
- This event has passed.
SBDI Days 2024: Towards Data-driven Ecology
January 24 @ 09:30 - January 25 @ 13:30 CET

The SBDI consortium welcomes everyone within the scientific community who is curious about data-driven research. Join us at the Swedish Museum of Natural History in Stockholm.
Ecology research has not yet reached the big data revolution. While the forthcoming ‘data deluge’ provides an amount of biodiversity and other environmental data not available before it also creates many challenges for researchers in fields of biodiversity, ecology, conservation biology and systematics.
With this two-day event we invite the Swedish biodiversity research community to come together and pave the way for data-driven research in ecology. It’s for everyone who wants to make use of the big data wave that is building up.
Symposium topics include big data research in macroecology, challenges for biodiversity data research and data driven ecology, machine-learning and artificial intelligence applications in conservation, perspectives on digital twins for ecosystems, and scaling up DNA-based ecology research.
SBDI Days: Towards data-driven ecology is hosted by the Swedish Biodiversity Data Infrastructure and will take place on January 24th-25th 2024 at the Swedish Museum of Natural History in Stockholm. The event will offer keynotes and presentations, posters, demonstrations, and time to mingle and exchange ideas about data driven ecology.
Registration: Deadline 2024-01-10. CLOSED
Presentations in addition to keynotes, poster or a demo: Deadline 2023-12-20. CLOSED
Language: English
Contact/Questions: Please contact support@biodiversitydata.se
Venue
Map, Public Transport, Parking, Lunch restaurant, Conference Dinner, Getting around:
See PDF with Full Program
Schedule (PDF with Full Program)
| Time | ||
| 24. January 2024 | ||
| 09.30 | Registration & coffee | |
| 10.00 | Welcome | |
| 10.30 | Symposium 1: Perils and promises in big data research. Keynotes and Talks | |
| 12.00 | Lunch | |
| 13.00 | Symposium 2: Machine-learning and artificial intelligence applications in conservation and management of ecosystems. Keynotes and Talks | |
| 14.30 | Coffee | |
| 15.00 | Workshops and Postersession | |
| 19.00 | Dinner | |
| 25. January 2024 | ||
| 09.00 | Symposium 3: Perspectives on digital twins for ecosystems and ecological modelling. Keynotes and Talks | |
| 10.30 | Coffee | |
| 11.00 | Symposium 4: Scaling up DNA-based ecology research. Keynotes and Talks | |
| 12.45 | Lunch | |
—— See Full Program and Abstracts. ——
.
Invited keynote speakers
Perils and promises in big data research

Alexandre Antonelli
Royal Botanic Gardens Kew
Alexandre is a biodiversity scientist studying the distribution, evolution, threats and sustainable uses of species and developing methods to speed up scientific discovery and innovation. Building on extensive fieldwork in the American tropics and integrating his findings with comprehensive datasets, his research has demonstrated how the evolution of landscapes and biodiversity are intrinsically linked, revealed the origins and processes underlying current patterns and synthesised knowledge across many groups of species and regions. Antonelli is the Director of Science at the Royal Botanic Gardens, Kew and Professor in Biodiversity and Systematics at the University of Gothenburg, among other appointments and distinctions.
Talk: Illuminating the world’s biodiversity darkspots – Delivering on the Kunming-Montreal Global Biodiversity Framework requires filling the most critical knowledge gaps. Here I will present a framework for prioritising regions around the world with highest chances of yielding new species to science as well as most new geographic records. I will discuss how new technological developments, from remote sensing to citizen science, environmental DNA and semi-automated inventories can (and should) be integrated with expert surveys and analysed under robust methods in order to properly identify the most important areas for biological conservation and restoration needed to halt biodiversity loss.
More information about Alex: https://www.kew.org/science/our-science/people/alexandre-antonelli

Rob James Boyd
UK Centre for Ecology & Hydrology
Rob is a quantitative ecologist and methodologist, whose main interests are in statistics and biodiversity monitoring. He works mostly on sampling theory and developing methods to assess, mitigate and communicate the risk of bias that is typical of biodiversity data.
Talk: Monitoring biodiversity with non-random samples. Rob will start by explaining why it is challenging to monitor biodiversity without random samples. He will then introduce some methods for assessing the extent of the problem and mitigating it to the extent that it is possible.
More information about Rob: https://www.ceh.ac.uk/fr/staff/robin-boyd
Machine-learning and artificial intelligence applications in conservation and management of ecosystems

Tobias Andermann
Department of Organismal Biology, Systematic Biology, Uppsala University
SciLifeLab and Wallenberg Data Driven Life Science Fellow
Tobias is a biodiversity researcher dedicated to providing data and computational tools for combating the global biodiversity crisis. His group, the Biodiversity Data Lab, is working on the intersection of molecular biology, spatial ecology, and machine learning, with the mission to provide a more comprehensive view on the distribution of biodiversity, including hidden diversity of inconspicuous and even undescribed species through the use of environmental DNA.
Talk: Biodiversity data requirements of AI tools for biodiversity-positive decision making – There is an ever-increasing need for developing standardized ways of quantifying the biodiversity value of a site. This need arises from international and local policies (e.g. the COP15 UN Biodiversity agreement) as well as from the independent investments of companies world-wide in developing biodiversity-positive profiles, motivated by their public perception. This momentum provides an unprecedented opportunity to efficiently protect biodiversity on a large scale to turn around the alarmingly negative biodiversity trends of the recent decades and centuries. While individual biodiversity assessments for specific sites can be done through targeted inventories, either through taxonomic experts or increasingly through the use of environmental DNA, we also need ways of scaling up such biodiversity evaluations to larger scales, covering large areas. AI models provide a unique tool to leverage complex biodiversity and remote sensing data to make predictions of biodiversity on large spatial scales, while at the same time providing high spatial resolution.
In this talk I will present some of these models from my ongoing research and discuss their utility, but also their current shortcomings and limitations. One of the main bottlenecks that we are currently facing is the quality of the biodiversity data that is currently available, which suffer from huge biases in their spatial and taxonomic coverage. Environmental DNA, collected on large spatial scales and made available through standardized platforms such as SBDI are a promising way forward to satisfy these data needs and overcome some of the biggest shortcomings.
More information about Tobias: https://www.biodiversity.se


Eva Lindberg & Langning Huo
Department of Forest Resource Management, Division of Forest Remote Sensing, Swedish University of Agricultural Sciences
Eva is developing new automatic methods to derive information from remotely sensed data, primarily airborne laser scanning data, relevant for vegetation and habitat analysis and mapping. Her work includes the technical aspects for data analysis, but also research about the related natural processes, for example the influence of canopy structure on the habitats of forest birds, insects, and lichens.
More information about Eva: https://www.slu.se/en/ew-cv/eva-lindberg/ and https://www.mistradigitalforest.se/en/news/promising-results-for-a-new-method-of-assessing-biodiversity
Langning is a researcher in forest remote sensing, specializing in developing remote sensing solutions for monitoring forest health and disturbance, from individual-tree scale to landscape scale. She has experience with analyzing indicators from remote sensing data on forest biodiversity and conservation values. Her work also includes developing remote sensing tools to early identify forest disturbances such as bark beetle infestations and drought damages.
More information about Langning: https://internt.slu.se/cv-originalen/langning-huo/
Talk: Biodiversity indicators from remote sensing laser data. Remote sensing can provide information to map vegetation states and changes for large areas. Recent developments in remote sensing has resulted in data with increasingly higher resolution, both spectrally, spatially, and temporally. This talk will give an introduction to mapping of biodiversity indicators from remote sensing and present how dead wood and trees with high conservation value can be mapped from high-resolution airborne laser scanning data.
Perspectives on digital twins for ecosystems and ecological modelling

Arne Jørgen Berre
SINTEF Digital, Smart Data group in Sustainable Communication Technologies department and NorwAI.ai
Arne is task lead of Digital Twins of the Ocean interoperability in the DTO-BioFlow project on bringing Biodiversity into the European Digital Twins of the Ocean. He is scientific coordinator in the Iliad Digital Twins of the Ocean project and further responsible for digital twins and EOSC interoperability in the AquaINFRA EOSC project and in the CLIMAREST Mission Ocean project.
He is co-lead in the European Big Data Value Association (BDVA.eu) task force on AI and Big Data standards and benchmarking, and involved in the ADRA organisation on AI, Data and Robotics. He is the lead of the Norwegian ISO SC42 AI group and involved in ISO SC41 IoT and Digital Twin, The Digital Twin Consortium, ISO/TC211 and OGC. He is co-chair of WG5 Digital Twins of the Ocean Framework and architecture (Turtle) in DITTO (Digital Twins of the Ocean) – a Global Program of the UN Decade of Ocean Science for Sustainable Development (2021-2030).
Talk: Ecosystem Digital Twin pipelines with Big Data and AI – Presents a standards-based framework for Ecosystem and Biodiversity digital twin pipelines, with examples from European projects like DTO-BioFlow, BioDT, Destination Earth/DestinE and Iliad/EDITO Digital Twins of the Ocean. A Digital Twin is a digital representation of a target entity with data connections that enable convergence between the physical and digital states at an appropriate rate of synchronization. Digital Twin pipelines are presented with a foundation in big data from new sensors and observations through multiple data types including observations, citizen science, video/images, remote sensing, bioacoustics, environmental DNA and others. Data management is supported through biodiversity data spaces and digital twin data lakes with the use of appropriate data representation standards and APIs and Digital Twin standards. Hybrid and Intelligent/Cognitive Digital Twins are combining data-driven models and simulation models with AI/Machine learning and natural models for predictions and recommendations supported by Cloud, GPU and HPC processing. Graphical digital twins offer various types of 2D/3D/4D visualisation and interaction modes.
Scaling up DNA-based ecology research

Monika Quinones Winder
Department of Ecology, Environment and Plant Sciences, Stockholm University
Monika is a professor in marine ecology at Stockholm University and a leader in aquatic food web interactions. By combining DNA metabarcoding, sustained monitoring datasets and network modelling, her group developed a novel approach to study the importance of diversity within community interactions. Current projects extend food web interactions from protists to fish building one of the most comprehensive and resolved marine food webs. She also explores the hidden role of zooplankton symbiotic interactions and impacts of shifting phenologies on trophic coupling. Monika earned a PhD from ETH, Switzerland and has worked at a number of international universities, including the University of Washington, University of Davis, USA and Geomar, Germany, before moving to Stockholm University.
Talk: Exploring ecological interactions with DNA-based network analysis – Food webs, composed of a vast diversity of organisms and their interactions are the engine of healthy, resilient and biodiverse coastal ecosystems. Yet, knowledge about organization of food webs is limited because of many methodological challenges to study ecological interactions in nature. Here I will present a new approach that combines DNA metabarcoding and energetic food web modelling to quantify carbon fluxes and ecosystem functioning in real and dynamic natural landscapes. This is supported by automated hydroacoustic data collection to investigate spatial distribution of plankton and fish in the water column in real time. Overall, these findings illustrate that an improved understanding of species interactions are key for endorsing essential measures to promote a resilient and biodiverse Baltic Sea, able to support socioeconomic activities.
Workshops
1. Research using camera traps

Summary: Research with camera traps. Automatic camera traps linked to AI-based image recognition are rapidly emerging power tools that provide repeatable and standardized sampling methods capable of expanding the extent and resolution of biodiversity monitoring. During the last few years, such traps in Sweden have begun providing data for moths, invasive alien plants and marine hard-bottom habitats. In this workshop we will showcase workflows, discuss the future of such data-driven, image-based approaches in ecology, and explore opportunities for future collaboration.
The workshop will 60 min and moderated by Lars B. Pettersson, Lund university.
2. Modelling the distribution of alien species in Sweden
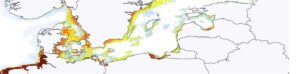
Summary: The number of alien species in terrestrial, marine and freshwater ecosystems is accumulating quickly and requires a systematic response and coordination between public authorities, the research community, and the private sector. Today new monitoring methods for alien species (e.g. eDNA, citizen science, etc) can be coupled with predictive analytical approaches such as Species distribution models to provide biological forecasts for distribution of alien species and invasive hotspots. Currently there are several initiatives which model invasive alien species in Sweden using new and conventional observation methods. The goal of this workshop is to bring these groups together and discuss links to the newly established Digital Twin projects BioDT, DTO-bioflow, and Illiad.
The workshop will be moderated by Matthias Obst, University of Gothenburg.
3. FAIR principles in life science research practice.

NBIS in collaboration with the SciLifeLab Data Centre.
Description: This workshop will showcase how to leverage the FAIR principles to guide effective data management practices for research groups and communities. It is specifically designed to cater to SBDI community members at all career stages and is organized by SciLifeLab Data Centre and NBIS – National Bioinformatics Infrastructure Sweden.
Instructors: Angela Fuentes Pardo, Arnold Kochari, Wolmar Nyberg Åkerström
4. Big data – big sample size? Assessing risk of bias
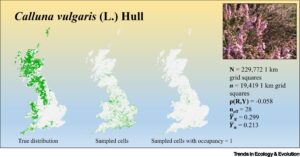
Summary: We will develop on the talk by Rob Boyd (Symposium 1: Perils and promises in big data research) and investigate deeper into why it is challenging to monitor biodiversity without random samples. We will assess risk of bias and investigate consequences for sample size and interpretation of results. See also We need to talk about nonprobability samples, ROBITT: A tool for assessing the risk-of-bias in studies of temporal trends in ecology, Descriptive inference using large, unrepresentative nonprobability samples: An introduction for ecologists, occAssess: An R package for assessing potential biases in species occurrence data.
Workshop held by Oliver Pescott and Rob James Boyd, UK Centre for Ecology & Hydrology.
Supporting materials for workshop 4:
- “occAssess” doc for assessing potential biases, example Swedish longhorn beetle (Cerambycidae) occurrence data at GBIF.org
- ROBITT tool doc, also available online as SI to the ROBITT paper, Appendix S1: Boyd Robin J. et al. , (2022). ROBITT: A tool for assessing the risk-of-bias in studies of temporal trends in ecology. Methods in Ecology and Evolution, 13, 1497–1507. https://doi.org/10.1111/2041-210X.13857
- occAssess paper: Boyd Robin J. et al. , (2021). occAssess: an R package for assessing potential biases in species occurrence data. Ecology and Evolution, 11, 16177-16187, http://dx.doi.org/10.1002/ece3.8299
